/ 9 min read
Deriving LSTM Gradient for Backpropagation
Recurrent Neural Network (RNN) is hot in these past years, especially with the boom of Deep Learning. Just like any deep neural network, RNN can be seen as a (very) deep neural network if we “unroll” the network with respect of the time step. Hence, with all the things that enable vanilla deep network, training RNN become more and more feasible too.
The most popular model for RNN right now is the LSTM (Long Short-Term Memory) network. For the background theory, there are a lot of amazing resources available in Andrej Karpathy’s blog and Chris Olah’s blog.
Using modern Deep Learning libraries like TensorFlow, Torch, or Theano nowadays, building an LSTM model would be a breeze as we don’t need to analytically derive the backpropagation step. However to understand the model better, it’s absolutely a good thing, albeit optional, to try to derive the LSTM net gradient and implement the backpropagation “manually”.
So, here, we will try to first implement the forward computation step according to the LSTM net formula, then we will try to derive the network gradient analytically. Finally, we will implement it using numpy.
LSTM Forward
We will follow this model for a single LSTM cell:
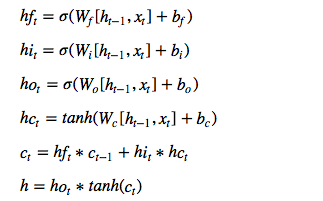
Let’s implement it!
import numpy as np
H = 128 # Number of LSTM layer's neuronsD = ... # Number of input dimension == number of items in vocabularyZ = H + D # Because we will concatenate LSTM state with the input
model = dict( Wf=np.random.randn(Z, H) / np.sqrt(Z / 2.), Wi=np.random.randn(Z, H) / np.sqrt(Z / 2.), Wc=np.random.randn(Z, H) / np.sqrt(Z / 2.), Wo=np.random.randn(Z, H) / np.sqrt(Z / 2.), Wy=np.random.randn(H, D) / np.sqrt(D / 2.), bf=np.zeros((1, H)), bi=np.zeros((1, H)), bc=np.zeros((1, H)), bo=np.zeros((1, H)), by=np.zeros((1, D)))Above, we’re declaring our LSTM net model. Notice that from the formula above, we’re concatenating the old hidden state h with current input x, hence the input for our LSTM net would be Z = H + D. And because our LSTM layer wants to output H neurons, each weight matrices’ size would be ZxH and each bias vectors’ size would be 1xH.
One difference is for Wy and by. This weight and bias would be used for fully connected layer, which would be fed to a softmax layer. The resulting output should be a probability distribution over all possible items in vocabulary, which would be the size of 1xD. Hence, Wy’s size must be HxD and by’s size must be 1xD.
def lstm_forward(X, state): m = model Wf, Wi, Wc, Wo, Wy = m['Wf'], m['Wi'], m['Wc'], m['Wo'], m['Wy'] bf, bi, bc, bo, by = m['bf'], m['bi'], m['bc'], m['bo'], m['by']
h_old, c_old = state
# One-hot encode X_one_hot = np.zeros(D) X_one_hot[X] = 1. X_one_hot = X_one_hot.reshape(1, -1)
# Concatenate old state with current input X = np.column_stack((h_old, X_one_hot))
hf = sigmoid(X @ Wf + bf) hi = sigmoid(X @ Wi + bi) ho = sigmoid(X @ Wo + bo) hc = tanh(X @ Wc + bc)
c = hf * c_old + hi * hc h = ho * tanh(c)
y = h @ Wy + by prob = softmax(y)
state = (h, c) # Cache the states of current h & c for next iter cache = ... # Add all intermediate variables to this cache
return prob, state, cacheThe above code is for the forward step for a single LSTM cell, which identically follows the formula above. The only additions are the one-hot encoding and the hidden-input concatenation process.
LSTM Backward
Now, we will dive into the main point of this post: LSTM backward computation. We will assume that derivative function for sigmoid and tanh are already known.
def lstm_backward(prob, y_train, d_next, cache): # Unpack the cache variable to get the intermediate variables used in forward step ... = cache dh_next, dc_next = d_next
# Softmax loss gradient dy = prob.copy() dy[1, y_train] -= 1.
# Hidden to output gradient dWy = h.T @ dy dby = dy # Note we're adding dh_next here dh = dy @ Wy.T + dh_next
# Gradient for ho in h = ho * tanh(c) dho = tanh(c) * dh dho = dsigmoid(ho) * dho
# Gradient for c in h = ho * tanh(c), note we're adding dc_next here dc = ho * dh * dtanh(c) dc = dc + dc_next
# Gradient for hf in c = hf * c_old + hi * hc dhf = c_old * dc dhf = dsigmoid(hf) * dhf
# Gradient for hi in c = hf * c_old + hi * hc dhi = hc * dc dhi = dsigmoid(hi) * dhi
# Gradient for hc in c = hf * c_old + hi * hc dhc = hi * dc dhc = dtanh(hc) * dhc
# Gate gradients, just a normal fully connected layer gradient dWf = X.T @ dhf dbf = dhf dXf = dhf @ Wf.T
dWi = X.T @ dhi dbi = dhi dXi = dhi @ Wi.T
dWo = X.T @ dho dbo = dho dXo = dho @ Wo.T
dWc = X.T @ dhc dbc = dhc dXc = dhc @ Wc.T
# As X was used in multiple gates, the gradient must be accumulated here dX = dXo + dXc + dXi + dXf # Split the concatenated X, so that we get our gradient of h_old dh_next = dX[:, :H] # Gradient for c_old in c = hf * c_old + hi * hc dc_next = hf * dc
grad = dict(Wf=dWf, Wi=dWi, Wc=dWc, Wo=dWo, Wy=dWy, bf=dbf, bi=dbi, bc=dbc, bo=dbo, by=dby) state = (dh_next, dc_next)
return grad, stateA bit long isn’t it? However, actually it’s easy enough to derive the LSTM gradients if you understand how to take a partial derivative of a function and how to do chain rule, albeit some tricky stuffs are going on here. For this, I would recommend CS231n.
Things that are tricky and not-so-obvious when deriving the LSTM gradients are:
- Adding
dh_nexttodh, becausehis branched in forward propagation: it was used iny = h @ Wy + byand the next time step, concatenated withx. Hence the gradient is split and has to be added here. - Adding
dc_nexttodc. Identical reason with above. - Adding
dX = dXo + dXc + dXi + dXf. Similar reason with above: X is used in many places so the gradient is split and need to be accumulated back. - Getting
dh_nextwhich is the gradient ofh_old. AsX = [h_old, x], thendh_nextis just a reverse concatenation: split operation ondX.
With the forward and backward computation implementations in hands, we could stitch them together to get a full training step that would be useful for optimization algorithms.
LSTM Training Step
This training step consists of three steps: forward computation, loss calculation, and backward computation.
def train_step(X_train, y_train, state): probs = [] caches = [] loss = 0. h, c = state
# Forward Step for x, y_true in zip(X_train, y_train): prob, state, cache = lstm_forward(x, state, train=True) loss += cross_entropy(prob, y_true)
# Store forward step result to be used in backward step probs.append(prob) caches.append(cache)
# The loss is the average cross entropy loss /= X_train.shape[0]
# Backward Step
# Gradient for dh_next and dc_next is zero for the last timestep d_next = (np.zeros_like(h), np.zeros_like(c)) grads = {k: np.zeros_like(v) for k, v in model.items()}
# Go backward from the last timestep to the first for prob, y_true, cache in reversed(list(zip(probs, y_train, caches))): grad, d_next = lstm_backward(prob, y_true, d_next, cache)
# Accumulate gradients from all timesteps for k in grads.keys(): grads[k] += grad[k]
return grads, loss, stateIn the full training step, first we’re do full forward propagation on all items in training set, then store the results which are the softmax probabilities and cache of each timestep into a list, because we are going to use it in backward step.
Next, at each timestep, we can calculate the cross entropy loss (because we’re using softmax). We then accumulate all of those loss in every timestep, then average them.
Lastly, we do backpropagation based on the forward step results. Notice while we’re iterating the data forward in forward step, we’re going the reverse direction here.
Also notice that dh_next and dc_next for the first timestep in backward step is zero. Why? This is because at the last timestep in forward propagation, h and c won’t be used in the next timestep, as there are no more timestep! So, the gradient of h and c in the last timestep are not split and could be derived directly without dh_next and dc_next.
With this function in hands, we could plug this to any optimization algorithm like RMSProp, Adam, etc with some modification. Namely, we have to take account on the state of the network. So, the state for the current timestep need to be passed to the next timestep.
And, that’s it. We can train our LSTM net now!
Test Result
Using Adam to optimize the network, here’s the result when I feed a copy-pasted text about Japan from Wikipedia. Each data is a character in the text. The target is the next character.
After each 100 iterations, the network are sampled.
It works like this:
- Do forward propagation and get the softmax distribution
- Sample the distribution
- Feed the sampled character as the input of next time step
- Repeat
And here’s the snippet of the results:
=========================================================================Iter-100 loss: 4.2125=========================================================================best c ehpnpgteHihcpf,M tt" ao tpo Teoe ep S4 Tt5.8"i neai neyoserpiila o rha aapkhMpl rlp pclf5i=========================================================================
...
=========================================================================Iter-52800 loss: 0.1233=========================================================================tary shoguns who ruled in the name of the Uprea wal motrko, the copulation of Japan is a sour the wa=========================================================================Our network definitely learned something here!
Conclusion
Here, we looked at the general formula for LSTM and implement the forward propagation step based on it, which is very straightforward to do.
Then, we derived the backward computation step. This step was also straightforward, but there were some tricky stuffs that we had to ponder about, especially the recurrency step in h and c.
We then stitched the forward and backward step together to build the full training step that can be used with any optimization algorithm.
Lastly, we tried to run the network using some test data and showed that the network was learning by looking at the loss value and the sample of text that are produced by the network.