/ 6 min read
Conditional Variational Autoencoder: Intuition and Implementation
Conditional Variational Autoencoder (CVAE) is an extension of Variational Autoencoder (VAE), a generative model that we have studied in the last post. We’ve seen that by formulating the problem of data generation as a bayesian model, we could optimize its variational lower bound to learn the model.
However, we have no control on the data generation process on VAE. This could be problematic if we want to generate some specific data. As an example, suppose we want to convert a unicode character to handwriting. In vanilla VAE, there is no way to generate the handwriting based on the character that the user inputted. Concretely, suppose the user inputted character ‘2’, how do we generate handwriting image that is a character ‘2’? We couldn’t.
Hence, CVAE [1] was developed. Whereas VAE essentially models latent variables and data directly, CVAE models lantent variables and data, both conditioned to some random variables.
Conditional Variational Autoencoder
Recall, on VAE, the objective is:
that is, we want to optimize the log likelihood of our data
Looking closely at the model, we could see why can’t VAE generate specific data, as per our example above. It’s because the encoder models the latent variable
Similarly, in the decoder part, it only models
We could improve VAE by conditioning the encoder and decoder to another thing(s). Let’s say that other thing is
Hence, our variational lower bound objective is now in this following form:
i.e. we just conditioned all of the distributions with a variable
Now, the real latent variable is distributed under
CVAE: Implementation
The conditional variable
Let’s use MNIST for example. We could use the label as our conditional variable
mnist = input_data.read_data_sets('MNIST_data', one_hot=True)X_train, y_train = mnist.train.images, mnist.train.labelsX_test, y_test = mnist.test.images, mnist.test.labels
m = 50n_x = X_train.shape[1]n_y = y_train.shape[1]n_z = 2n_epoch = 20
# Q(z|X,y) -- encoderX = Input(batch_shape=(m, n_x))cond = Input(batch_shape=(m, n_y))The natural question to arise is how do we incorporate the new conditional variable into our existing neural net? Well, let’s do the simplest thing: concatenation.
inputs = merge([X, cond], mode='concat', concat_axis=1)
h_q = Dense(512, activation='relu')(inputs)mu = Dense(n_z, activation='linear')(h_q)log_sigma = Dense(n_z, activation='linear')(h_q)Similarly, the decoder is also concatenated with the conditional vector:
def sample_z(args): mu, log_sigma = args eps = K.random_normal(shape=(m, n_z), mean=0., std=1.) return mu + K.exp(log_sigma / 2) * eps
# Sample z ~ Q(z|X,y)z = Lambda(sample_z)([mu, log_sigma])z_cond = merge([z, cond], mode='concat', concat_axis=1) # <--- NEW!
# P(X|z,y) -- decoderdecoder_hidden = Dense(512, activation='relu')decoder_out = Dense(784, activation='sigmoid')
h_p = decoder_hidden(z_cond)outputs = decoder_out(h_p)The rest is similar to VAE. Heck, even we don’t need to modify the objective. Everything is already expressed in our neural net models.
def vae_loss(y_true, y_pred): """ Calculate loss = reconstruction loss + KL loss for each data in minibatch """ # E[log P(X|z,y)] recon = K.sum(K.binary_crossentropy(y_pred, y_true), axis=1) # D_KL(Q(z|X,y) || P(z|X)); calculate in closed form as both dist. are Gaussian kl = 0.5 * K.sum(K.exp(log_sigma) + K.square(mu) - 1. - log_sigma, axis=1)
return recon + klFor the full explanation of the code, please refer to my original VAE post. The full code could be found in my Github repo: https://github.com/wiseodd/generative-models.
Conditional MNIST
We will test our CVAE model to generate MNIST data, conditioned to its label. With the above model, we could specify which digit we want to generate, as it is conditioned to the label!
First thing first, let’s visualize
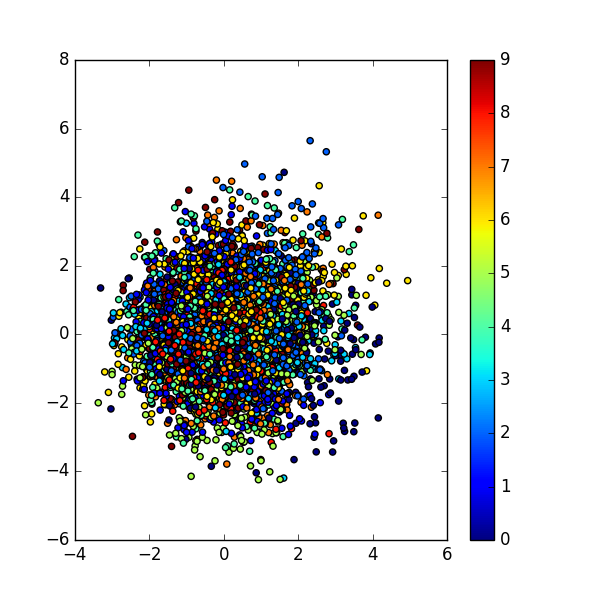
Things are messy here, in contrast to VAE’s
Next, let’s try to reconstruct some images:
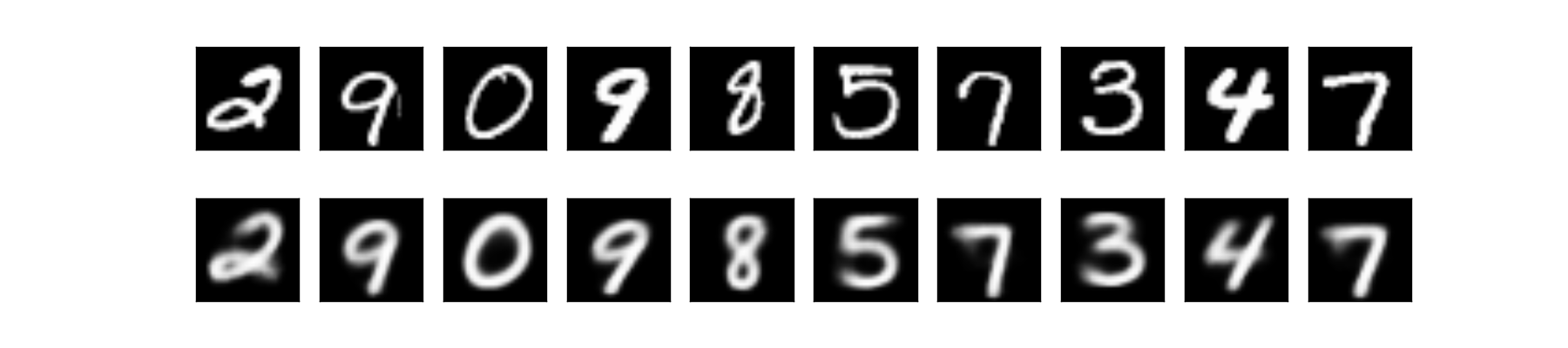
Subjectively, we could say the reconstruction results are way better than the original VAE! We could argue that because each data under specific label has its own distribution, hence it is easy to sample data with a specific label. If we look back at the result of the original VAE, the reconstructions suffer at the edge cases, e.g. when the model is not sure if it’s 3, 8, or 5, as they look very similar. No such problem here!
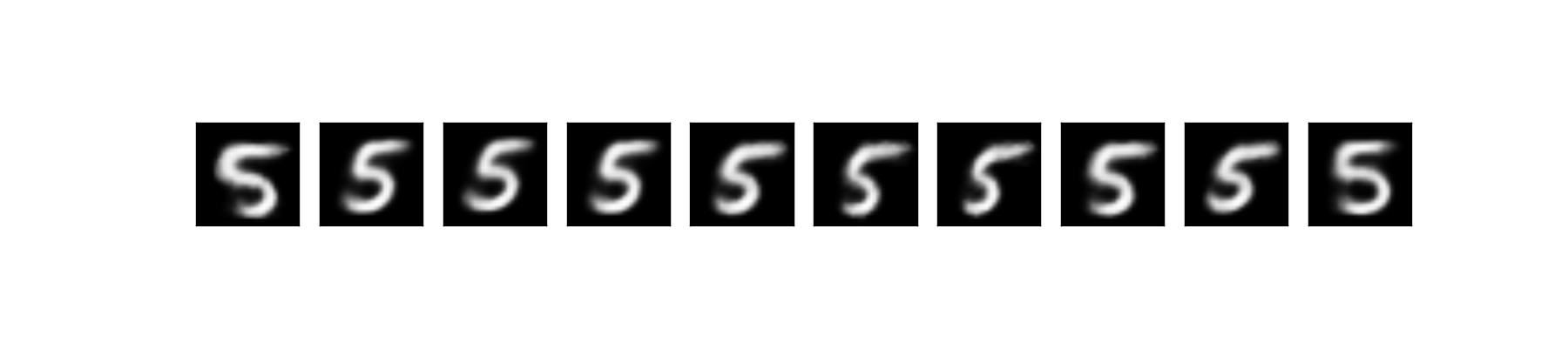
Now the interesting part. We could generate a new data under our specific condition. Above, for example, we generate new data which has the label of ‘5’, i.e.
Conclusion
In this post, we looked at the extension of VAE, the Conditional VAE (CVAE).
In CVAE, we could generate data with specific attribute, an operation that can’t be done with the vanilla VAE. We showed this by applying CVAE on MNIST data and conditioned the model to the images’ labels. The resulting model allows us to sample data under specific label.
We also noticed that by conditioning our MNIST data to their labels, the reconstruction results is much better than the vanilla VAE’s. Hence, it is a good thing, to incorporate labels to VAE, if available.
Finally, CVAE could be conditioned to anything we want, which could result on many interesting applications, e.g. image inpainting.
References
- Sohn, Kihyuk, Honglak Lee, and Xinchen Yan. “Learning Structured Output Representation using Deep Conditional Generative Models.” Advances in Neural Information Processing Systems. 2015.